An Investigation of the Sources of Fecal Contamination
at Petrie Island Beach on the Ottawa River in 2007
Thomas A. Edge, Stephen Hill and Flora Suen
National Water Research Institute
Water Science & Technology Directorate
Environment Canada
Burlington, Ontario
February 22, 2008
A study was conducted by the National Water Research Institute, Environment Canada, to investigate the source of fecal pollution responsible for beach postings at Petrie Island Beach near the City of Ottawa in 2007. Field observations and water samples were collected weekly at the Beach and at upstream Ottawa River locations from May 7 to October 15, 2007. Water samples were analyzed for E. coli, and detection of a DNA marker for anaerobic Bacteroides bacteria associated with human fecal pollution.
Field observations found evidence of fecal contamination at Petrie Island Beach from both bird and human fecal sources. Bird droppings were numerous at some places, although their role in beach postings or human health risks remains uncertain. Observation of floatables on the beach at times, was indicative of the impacts of human fecal pollution. E. coli concentrations at Petrie Island Beach were higher in the sand than in the adjacent beach water, and they increased in the sand and beach water over the course of the bathing season. These results are consistent with other studies that have found sand serving as a reservoir for E. coli. The human Bacteroides DNA marker was detected in 26% of the beach water samples from Petrie Island Beach in 2007. Both E. coli concentrations and the occurrence of the human Bacteroides DNA marker were much numerous at the Beach associated with rain events than during dry weather.
Water quality monitoring upstream of Petrie Island Beach found that E. coli concentrations and the occurrence of the human Bacteroides DNA marker were lowest at the Ottawa River transect above the City of Ottawa and City of Gatineau municipal wastewater treatment plant outfalls. At each transect, E. coli concentrations and the human Bacteroides marker were more numerous at sampling locations on the Quebec side of the Ottawa River than on the Ontario side. An exception was the higher occurrence of the human Bacteroides DNA marker at the sampling location just downstream from the outfall of the City of Ottawa wastewater treatment plant. Bilberry Creek had the highest E. coli concentrations and occurrence of the human Bacteroides DNA marker among three small tributaries to the Ottawa River.
The impacts of human fecal pollution at Petrie Island Beach were sporadic in 2007, and mostly associated with rain events. It will be important to better understand when these impacts are likely to occur in the future, and at a minimum, a no swimming rain rule would seem appropriate. The source of human fecal contamination at the Beach appeared to be most closely associated with fecal pollution sources on the Ontario side of the Ottawa River in 2007. It will be important to monitor water quality in the future at Bilberry Creek and downstream of the outfall of the City of Ottawa wastewater treatment plant (ROPEC). Close communication between operators of the ROPEC wastewater treatment plant and public health officials overseeing Petrie Island Beach would be needed. It is important to recognize that there are limitations to using a bacterial indicator like E. coli for making water quality decisions in a riverine setting downstream from a chlorinated outfall of a large municipal wastewater treatment plant. Additional research would be required to better define and remediate the human fecal pollution sources having the most significant impact on Petrie Island Beach.
Table of Contents
Executive Summary (French Translation)
Field Observations at Petrie Island Beach
Water Quality at the Petrie Island Beach
Occurrence of Human Bacteroides DNA marker
Water Quality upstream in the Ottawa River
and tributaries
Occurrence of Human Bacteroides DNA marker
Petrie Island Beach is located on the south shore of the Ottawa River about 20 km east of the City of Ottawa. The beach was established in 2005, and it is the only recreational beach in the east end of Ottawa. The City of Ottawa conducted a two-year water quality monitoring study for the beach in 2003-04, and began regular water quality monitoring in 2005 (City of Ottawa, 2005). This study found microbial water quality to be generally good at the beach in 2003-05 with seasonal geometric mean E. coli concentrations ranging from 37 to 74 CFU/100mL. However, high concentrations of E. coli were found at the beach at times in 2006 raising questions about the source of fecal contamination responsible for beach postings.
There are a variety of potential sources of fecal contamination that could impact Petrie Island Beach ranging from distant wastewater inputs into the Ottawa River upstream of the beach to localized bird dropping inputs right at the beach. The Cities of Ottawa, Ont. and Gatineau, Que. occur upstream of Petrie Island Beach and are a potential source of fecal contamination into the Ottawa River from stormwater runoff and combined sewer outfalls. Also upstream, the Rideau and Gatineau Rivers flow into the Ottawa River, and there are smaller tributaries immediately upstream of the beach such as Green Creek and Bilberry Creek. In addition, sewage treatment plants for the Cities of Ottawa and Gatineau discharge into the Ottawa River upstream of Petrie Island Beach. It is also known that birds such as gulls and Canada geese occur at Petrie Island Beach, and their fecal droppings could contribute to loadings of E. coli at the beach. While municipal wastewater is a familiar source of E. coli contamination at many beaches across Canada, recent studies at beaches around the Great Lakes have also drawn attention to the importance of more localized sources of E. coli at beaches such as from bird droppings (Edge and Hill, 2007), resuspension of sand (Whitman and Nevers, 2003), or runoff from impervious surfaces such as parking lots (Scopel et al. 2006; Edge et al., 2007).
The following study applied field observations, E. coli enumeration, and a microbial source tracking approach to investigate the potential sources of fecal contamination at Petrie Island Beach in 2007. Microbial source tracking techniques compare the similarity of microorganisms from fecal pollution sources and water samples in order to make inferences about the source of water contamination (U.S. EPA, 2005; Edge and Schaefer, 2006). There are two general approaches to microbial source tracking: library-dependent methods and library-independent methods. Library-dependent methods select an indicator microorganism like E. coli, and collect hundreds to thousands of isolates from fecal sources and water samples of interest. The similarity of fecal and water E. coli isolates is measured by DNA fingerprinting or other forensic-like techniques to infer the likely source of the water isolates. While these methods can provide very useful information for beach managers (Edge and Hill, 2007), they can be relatively expensive and time consuming. Given some of these considerations, a library-independent method was selected to begin investigation of Petrie Island Beach.
Library-independent source tracking methods are based on searching for host-specific microorganisms in water samples rather than building large libraries of fecal indicator bacteria. These host-specific microorganisms are adapted to specific gastrointestinal tracts, and they occur only in the feces of their host such as humans or ruminant animals. If the DNA sequence of such a microorganism is detected in a water sample, it is an indication of fecal contamination from that host human or animal. Some of the most promising library-independent methods are based on detecting host-specific strains of the anaerobic bacterium Bacteroides. This bacterium is generally found in much greater numbers in gastrointestinal tracts than E. coli. In addition, human-specific strains of Bacteroides are increasingly under investigation as indicators of the presence of fecal contamination from sources like municipal wastewaters (Field and Samadpour, 2007). The present study sought to determine the frequency of a human Bacteroides DNA sequence in water samples from Petrie Island Beach and upstream locations.
Water sampling at Petrie Island Beach and upstream locations was conducted by the City of Ottawa between May 7 and October 15, 2007. The sampling was conducted on a weekly basis during the bathing season. The sampling locations for Petrie Island Beach, upstream transects across the Ottawa River, and several upstream tributaries are shown in Figure 1. On each water sampling day, observations were made of the number of floatables (e.g. tampon applicators), birds, and bird fecal droppings at Petrie Island Beach. Floatables and bird droppings were enumerated along the beach within about 2 meters of the water’s edge. Data on rainfall from the rain gauge at ROPEC municipal wastewater treatment plant was provided by the City of Ottawa. A rain event water sample for the purposes of this study was defined as one where there was a cumulative rainfall of greater than 5 mm in the 24 hours before water sample collection.
Water samples were collected in sterile
500mL bottles and shipped overnight on ice to the National Water Research
Institute, Environment Canada, in Burlington ON. Sand pore water samples
at the beach were collected by digging into the sand at the edge of the water
and allowing pore water to seep into the hole. E.
coli concentrations in water
(CFU/100mL) were determined by filtering water samples through 0.45
µm filters (Millipore) and incubating the filters on
chromogenic differential coliform (DC) media (Oxoid Inc.) at 44.5°C for 18 hrs.
Water samples were also screened for the presence of strains of the anaerobic
bacterium Bacteroides that are associated with human fecal pollution
(Bernhard and Field, 2000; Bower et al., 2005). The
latter assay involved filtering as much water as the sample permitted,
up to 300 mL (50mL for sand pore water), and
extracting total genomic DNA from the filter. Filters were frozen at
-80°C before DNA extraction. The filter was first homogenized in a
Mini-Beadbeater (BioSpec Products Inc.) for 2 min. DNA
was purified using a Powersoil DNA isolation
kit (Mo BIO Laboratories, Inc.). A 1 µl extract was
used as template in a polymerase chain reaction (PCR)
assay using primer HF183F to amplify the human Bacteroides DNA sequences
and BAC32 to amplify generic Bacteroides
sequences if they were present in the sample. Primer BAC708R was the
reverse primer for both reactions. For the PCR reaction, the following
concentrations were used: 0.05 U/µL Hotmaster
Taq and 1 x buffer (Intermedico), 0.8 mM dNTP mixture, 0.06% BSA, 1.56 pmol/ µL each primer and water to 25 µL. The PCR cycling conditions were: 2 min at 94°C followed by 35 cycles of 20 sec at 94°C, 10 sec anneal at 53°C
for BAC32 or 63°C for HF183 primers, 50 sec at
65°C and a final single step at 65 °C for 7 min. A human fecal DNA extract was run as a
positive control for each set of reactions, along with sterile water as a
negative control. A 5 µL amount of dye DNA mix was loaded into wells of a 1.25%
agarose gel, and run at 170 V for approximately 1 hr to resolve the bands which
were visualized by staining with ethidium bromide and imaging under UV light.
Fecal samples were also collected in the Ottawa area over the course of the study period. DNA extracts from these fecal samples were used to test the specificity of the human Bacteroides source tracking method. Raw sewage samples were collected each sampling week from the influent of the City of Ottawa municipal wastewater treatment plant (the R.O. Pickard Environmental Centre - ROPEC). Fecal droppings from Canada geese and seagulls were collected along the Ottawa River. Fecal droppings from stray rabbits, cats and dogs were collected at the Ottawa Humane Society facilities on Champagne Avenue.
Statistical analyses were performed to explore whether the occurrence
of E. coli and human Bacteroides DNA marker at Petrie Island
Beach sampling locations was more similar to upstream sampling locations on the
Ontario or Quebec side of the Ottawa River.
Nonparametric spearman rank correlations were
performed to provide a general comparison of similarities between Petrie Island
Beach and upstream sampling locations for the occurrence of the human Bacteroides DNA marker and E. coli concentrations. A cluster analysis of Beach and upstream
sampling locations was performed by SAS JMP ver. 7 hierarchical clustering using
Ward’s method.
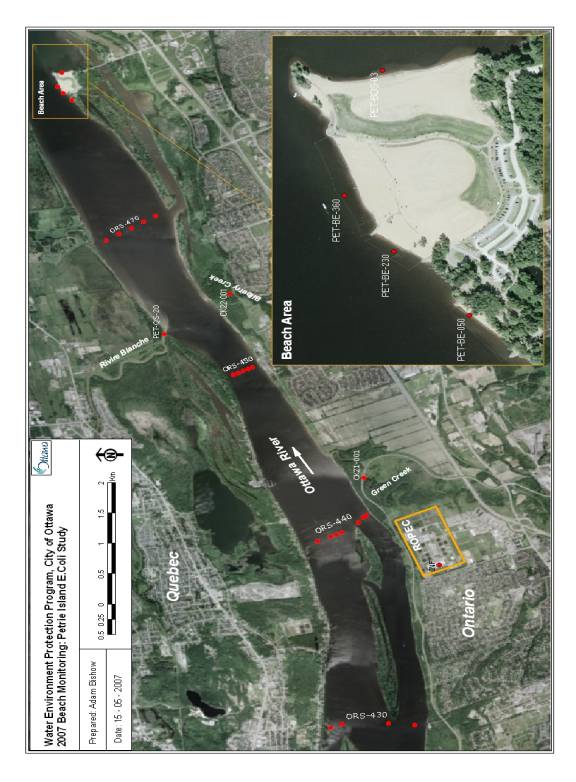

Figure 1. Petrie Island Beach study area showing sampling locations at the beach, and at upstream locations in the Ottawa River and tributaries.
Field Observations at
Petrie Island Beach
Petrie Island Beach was visited on April 27, 2007, before beach grooming and preparations for the 2007 bathing season had begun. There were clear indications of the impact of high water levels on the beach from spring flooding. Mats of floating debris had washed up on the beach, and there was evidence of contamination from untreated municipal wastewater. A walk along the entire length of the beach resulted in observations of 37 tampon applicators, 5 condoms, and 2 syringes. The numbers of these floatables counted over the 2007 bathing season is shown in Figure 2. These observations suggest there was some impact from untreated municipal wastewater at times in 2007 at Petrie Island Beach.
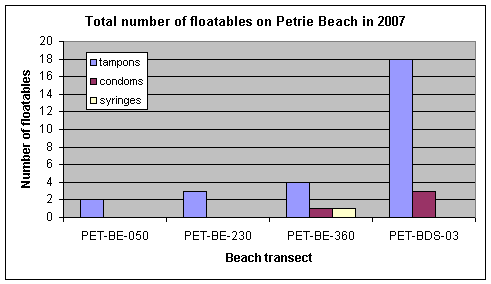
Figure 2. Total number of floatables counted from weekly observations along stretches of Petrie Island Beach near each sampling transect in 2007.
The total number of birds counted at Petrie Island from weekly observations in 2007 is presented in Figure 3. Gulls and Canada geese were observed on the beach, and they were more common at transects PET-BE-360 and PET-BDS-03 (Baie des Sables). The numbers of birds observed at Petrie Island Beach was lower than observed at some beaches along Lake Ontario where there can be over two hundred gulls and Canada geese observed at a beach on one day.
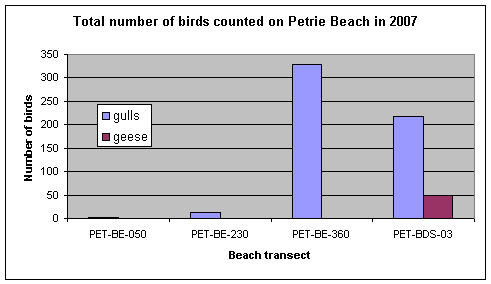
Figure 3. Total number of birds counted from weekly observations at Petrie Island Beach near each sampling transect in 2007.
The cumulative number of bird droppings counted within 2 meters of the waterline along Petrie Island Beach in 2007 is shown in Figure 4. Most of the bird droppings were from gulls, and they were largely deposited on the sand along the beach near the Baie des Sables transect. Individual gulls can produce between 34 and 62 fecal droppings in 24 hours (Gould and Fletcher, 1978), and E. coli numbers in gull feces have been reported as high as 1.9 x 109 CFU/gram of feces (Fogarty et al. 2003). While these bird droppings would have provided some level of continuous loading of E. coli into the beach sand at Petrie Island Beach, their significance for beach postings in 2007 is uncertain. For comparison, there is a beach about the length of the PET-BDS-03 area in Hamilton Harbour that is significantly impaired by bird droppings. This beach had cumulative numbers of more than 3000 gull and 3000 geese droppings over a bathing season, and poor water circulation to flush nearshore beach water (Edge and Hill, 2007).
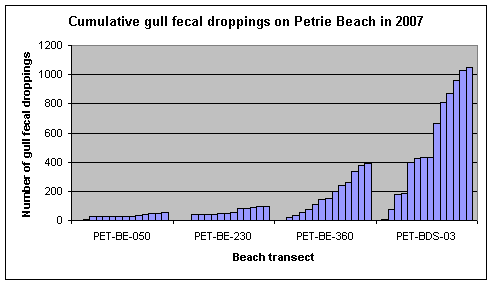
Figure 4. Cumulative number of gull fecal droppings counted from weekly observations along stretches of Petrie Island Beach near each sampling transect in 2007.
Water Quality at the
Petrie Island Beach
E. coli concentrations at Petrie Island Beach were found to be much higher in sand (pore water) than in the adjacent beach water (Table 1). They were also found to increase over the study period, except for a decline in E. coli numbers in the sand in the fall months. These results are consistent with many other studies that have found that sand can serve as a reservoir for accumulating E. coli over the course of a bathing season (Whitman and Nevers, 2003; Edge and Hill, 2007). Concentrations of E. coli in beach water were occasionally high, reaching as high as 33,000 CFU/100mL in ankle depth water and 1760 CFU/100mL in chest depth water in 2007.
Table 1. Monthly geometric mean E. coli concentration (CFU/100mL) in sand (pore water) and water depth zones (ankle, chest) at four Petrie Island Beach transects in 2007.
|
Location |
May (n=12) |
June (n=20) |
July (n=16) |
August (n=16) |
Sep/Oct (n=8) |
|
Pore water |
283 |
3920 |
4621 |
5157 |
3541 |
|
Ankle depth |
22 |
70 |
177 |
183 |
297 |
|
Chest depth |
16 |
43 |
66 |
65 |
211 |
A summary of the variability of E. coli concentrations at each transect along Petrie Island Beach is shown in Figure 5. At each transect, E. coli concentrations are much higher in the sand pore water than adjacent beach water. There was a tendency for E. coli concentrations to be highest in the sand pore water at the Baie des Sables transect (PET-BDS-03-P). This might be related to an increased loading from gull droppings there.
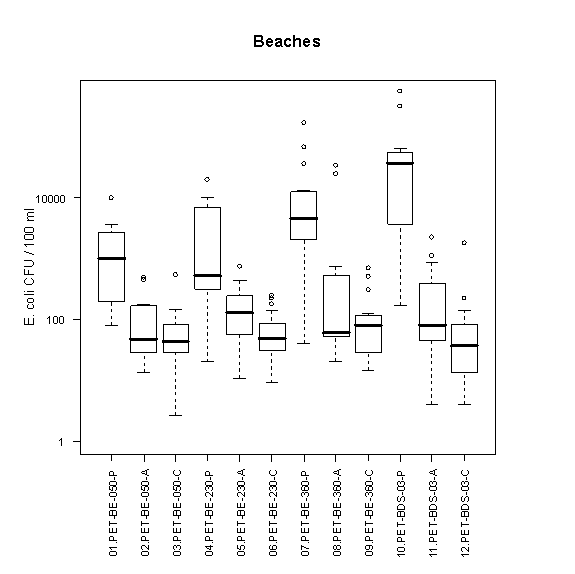
Figure 5. Box plots showing median and range of E. coli concentrations (CFU/100mL) for different transects and sampling locations at Petrie Island Beach in 2007. Results are presented for each transect identified in Figure 1 for water samples collected in sand pore water (e.g. location PET-BDS-050-P), ankle depth water (e.g. location PET-BDS-050-A), and chest depth water (e.g. location PET-BDS-050-C).
E. coli concentrations were found to be higher at Petrie Island Beach associated with rain events than during dry weather (Table 2). This is similar to the situation at many other beaches where rain events are known to result in increased levels of indicator bacteria like E. coli from a variety of sources such as surface water run-off from land, stormwater and combined sewer overflows, and sediment resuspension. While there were only four rain event sampling days in 2007, geometric mean E. coli concentrations for these rain days were twice as high as during dry weather at Petrie Island Beach.
Table 2. Geometric mean E. coli concentrations (CFU/100mL) during rain event and dry weather sampling days at four transects at Petrie Island Beach in 2007. There were four rain sampling days providing 16 water samples.
|
Location |
Rain sampling days (n=16) |
Dry sampling days (n=51) |
|
Pore water |
4363 |
2143 |
|
Ankle depth |
219 |
65 |
|
Chest depth |
76 |
37 |
Occurrence of Human
Bacteroides DNA marker
An evaluation of the specificity of the human Bacteroides DNA marker is found in Table 3. This marker was readily detected in most of the samples of raw sewage from the ROPEC wastewater treatment plant. However, it was never detected in any of the fecal samples from gulls, Canada geese, rabbits, cats, or dogs. It was also never detected in samples of sterile water used as negative controls in our laboratory. These results are consistent with other microbial source tracking studies that have found this human Bacteroides DNA marker to be a useful indicator of human fecal waste (Bernhard and Field, 2000; Bower et al., 2005; Field and Samadpour, 2007).
Table 3. Occurrence of the human Bacteroides DNA marker in fecal samples collected from the Ottawa area during the 2007 field season.
|
Fecal source |
No. of fecal samples |
% +ve for human Bacteroides |
|
Seagull dropping |
7 |
0 |
|
Canada geese dropping |
14 |
0 |
|
Rabbit dropping |
1 |
0 |
|
Cat dropping |
11 |
0 |
|
Dog dropping |
12 |
0 |
|
ROPEC sewage |
18 |
78 |
The human Bacteroides DNA marker was detected in 26% of ankle and chest depth water samples collected at Petrie Island Beach in 2007 (n=141 water samples). The DNA marker was detectable each month at the beach over the course of the 2007 study period (Table 4). It was most commonly detected in May and the fall months outside of the bathing season. Unlike E. coli, this Bacteroides DNA marker was less commonly detected in the sand pore water than adjacent beach water. It is possible that the Bacteroides bacteria do not survive and persist in beach sand as well as E. coli. It is also possible that the smaller sample volume (50mL) used for pore water, or contaminants in the sand pore water interfering with the PCR assay may have resulted in false negative results for the human Bacteroides DNA marker. We do not think the latter was common because amplification of the generic Bacteroides DNA marker was usually successful in almost all water samples. We also do not think there is evidence for much concern about false positive results for the human Bacteroides DNA marker. This DNA marker is being used successfully in an increasing number of microbial source tracking studies around the world (Gawler et al. 2007). However, to provide additional rigor, we will sequence some PCR amplicons from water samples presumed to be the human Bacteroides DNA marker.
Table 4. Percent of water samples that were positive for the human Bacteroides DNA marker in sand (pore water) and at different depth zones (ankle and chest) at four Petrie Island Beach transects in 2007.
|
Location |
May (n=12) |
June (n=20) |
July (n=16) |
August (n=16) |
Sep/Oct (n=8) |
|
Pore water |
8 |
0 |
6 |
0 |
38 |
|
Ankle depth |
42 |
22 |
6 |
6 |
100 |
|
Chest depth |
42 |
15 |
19 |
0 |
100 |
The frequency of occurrence of the human Bacteroides DNA marker at each transect at Petrie Island Beach is shown in Figure 6. This marker was less frequent in the sand than adjacent beach water at each transect, and it was never detected in pore water at the Baie des Sables transect. The marker was usually detectable in a least 20% of beach water samples at each transect over the 2007 sampling period.
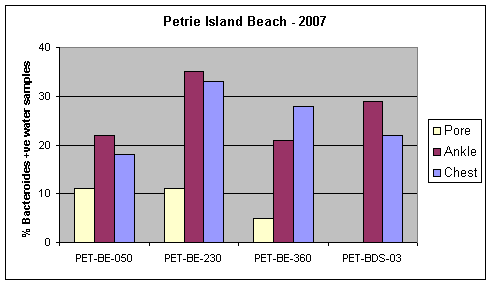
Figure 6. Occurrence of the human Bacteroides DNA marker in water samples collected at the four transects at Petrie Island Beach in 2007.
The human Bacteroides DNA marker was detected more frequently at Petrie Island Beach during rain events than dry weather in 2007 (Table 5). While the marker was never common during dry weather, it was detected in over 50% of beach water samples collected at ankle and chest depth on rain event days. The number of rain event days in 2007 was small (4 days) and included some relatively minor rain days when the cumulative rainfall over the previous 24 hours was 12.6mm (May 28), 6.2mm (June 28), 33.4mm (July 9), and 7.8mm (August 7). For example, there were no human Bacteroides DNA marker detections in beach water samples for the August 7 rain event. However, for the May 28 and June 28 rain event days, the human Bacteroides DNA marker was found to be widespread in beach water samples. This was also the case for the two fall sampling days.
Table 5. Percent of water samples that were positive for the human Bacteroides DNA marker during rain event and dry weather sampling days at four transects at Petrie Island Beach in 2007. There were four rain sampling days providing 16 water samples.
|
Location |
Rain sampling days (n=16) |
Dry sampling days (n=51) |
All sampling days (n=67) |
|
Pore water |
6 |
2 |
3 |
|
Ankle depth |
56 |
4 |
17 |
|
Chest depth |
56 |
4 |
17 |
The human Bacteroides DNA marker was more common in chest depth water samples when E. coli concentrations were high (Figure 7). Only one water sample was negative for the human Bacteroides DNA marker when E. coli levels were > 300 CFU/100mL. This was a sample collected on August 27 when the E. coli concentration was 500 CFU/100/mL. Although not as common, the human Bacteroides DNA marker was detected at times when E. coli concentrations were low. Water samples collected on May 28 and July 4 had the DNA marker present when E. coli concentrations were only 9 CFU/100mL. These and other results indicate that there can be human fecal contamination present, even though E. coli concentrations are below the recreational guideline level of 100 CFU/100mL.
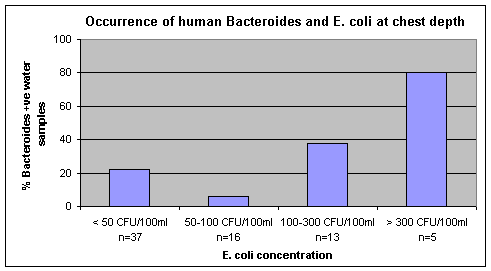
Figure 7. Occurrence of the human Bacteroides DNA marker in chest depth water samples with different levels of E. coli at Petrie Island Beach in 2007.
Water Quality upstream in
the Ottawa River and tributaries
Water quality monitoring upstream of Petrie Island Beach found the lowest geometric mean E. coli concentrations at Ottawa River transect 430 above the City of Ottawa and City of Gatineau municipal wastewater treatment plant outfalls (Figure 8). Higher geometric mean E. coli concentrations were found at transect 450 below these outfalls. The highest E. coli concentration measured at this transect was 8900 CFU/100mL on October 15. Bilberry Creek had the highest geometric mean concentration (615 CFU/100mL) among the tributaries to the Ottawa River. E. coli concentrations were high in this Creek during rain events and reached 7100 CFU/100mL on July 9.
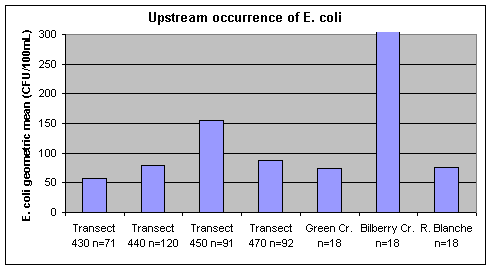
Figure 8. Geometric mean concentrations of E. coli at Ottawa River transects and tributaries upstream of Petrie Island Beach in 2007. The geometric mean concentration for E. coli in Bilberry Creek was 615 CFU/100mL.
A summary of the variability of E. coli concentrations at each upstream transect across the Ottawa River is shown in Figure 9. At each transect, E. coli concentrations were higher at sampling locations on the Quebec side of the Ottawa River. The highest E. coli concentration measured on the Quebec side was 9100 CFU/100mL at station 440.70 (below Gatineau wastewater plant outfall) on October 15. The highest E. coli concentration on the Ontario side was 1400 CFU/100mL at station 440.20 (below Ottawa wastewater plant outfall) on August 7.
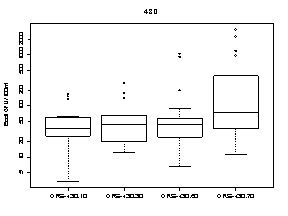
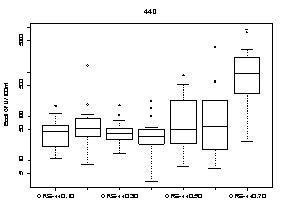
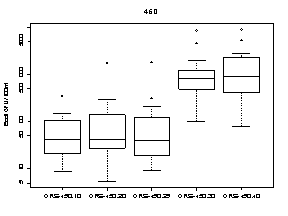
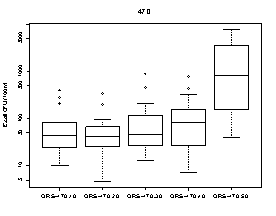
Figure 9. Box plots showing median and range of E. coli concentrations (CFU/100mL) for sampling locations on four different transects across the Ottawa River upstream of Petrie Island Beach in 2007. Results are presented for each transect showing sampling locations progressing from the Ontario side to the Quebec side of the River.
Upstream E. coli concentrations in the Ottawa River and tributaries were found to be higher associated with rain events than during dry weather (Figure 10). This is similar to the situation for many water bodies where rain events are known to result in increased levels of indicator bacteria like E. coli from a variety of sources such as surface water run-off from land, stormwater and combined sewer overflows, and sediment resuspension. While there were only four rain event days in 2007, geometric mean E. coli concentrations for these rain days were over twice those for dry weather days at transects downstream of the wastewater treatment plant outfalls and in Bilberry Creek.
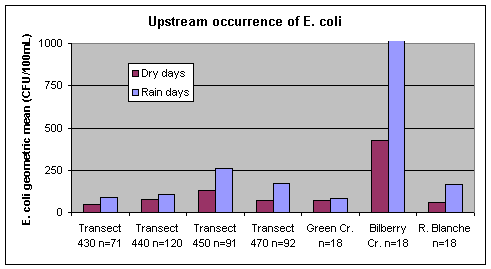
Figure 10. Geometric mean concentrations of E. coli for dry and rain sampling days at Ottawa River transects and tributaries upstream of Petrie Island Beach in 2007. The geometric mean for E. coli on rain days in Bilberry Creek was 2190 CFU/100mL.
Occurrence of Human
Bacteroides DNA marker
Water quality monitoring in the Ottawa River upstream of Petrie Island Beach found the lowest frequency of occurrence of the human Bacteroides DNA marker at transect 430 above the City of Ottawa and City of Gatineau municipal wastewater treatment plant outfalls (Figure 11). The occurrence of this DNA marker was more frequent at transect 450 below these outfalls. The highest frequency of occurrence of the human Bacteroides DNA marker was found in Bilberry Creek.
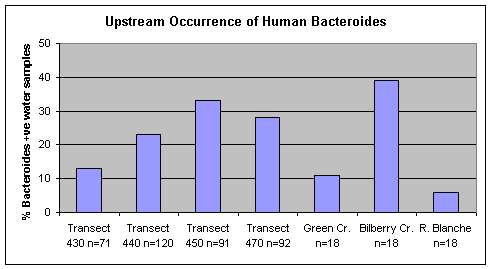
Figure 11. Occurrence of the human Bacteroides DNA marker in water samples collected at Ottawa River transects and tributaries upstream of Petrie Island Beach in 2007.
The highest frequency of occurrence of the human Bacteroides DNA marker was generally found along the Quebec shore of the Ottawa River (Figure 12). The one exception was the sampling location downstream of the City of Ottawa wastewater treatment plant outfall (440.2).
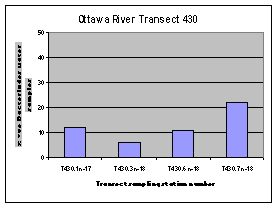
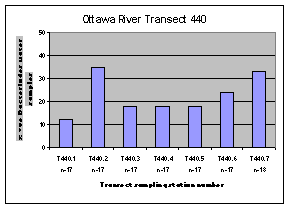
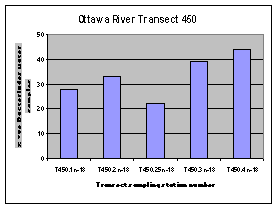
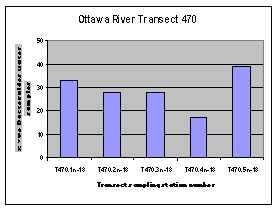
Figure 12. Occurrence of the human Bacteroides DNA maker at sampling locations on four Ottawa River transects upstream of Petrie Island Beach in 2007.
The human Bacteroides DNA marker was detected more frequently during rain events than dry weather at Ottawa River transects upstream of Petrie Island Beach in 2007 (Figure 13). The DNA marker was only detected twice at sampling station 430.10 on the Ontario side upstream of the municipal wastewater treatment plant, and these were both rain event sampling days (May 28 and June 28). While the DNA marker was more frequent in Bilberry Creek on rain sampling days, this was not the case for the other Creeks. These results indicate that during rain events there is more frequent detection of human fecal matter in the Ottawa River and Bilberry Creek.
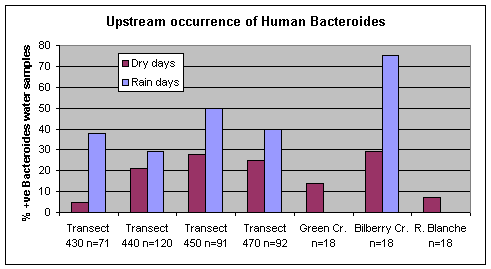
Figure 13. Percent of water samples that were positive for the human Bacteroides DNA marker during rain event and dry weather sampling days at Ottawa River transects and tributaries upstream of Petrie Island Beach in 2007.
Spearman rank correlation analyses found that the occurrence of the
human Bacteroides DNA marker at
Petrie Island Beach was most similar to its occurrence at the sampling location
440.2 just below the City of Ottawa municipal wastewater outfall (R2
= 0.88). The next highest spearman correlation coefficient was for sampling
site 470.1 just upstream of the beach on the Ontario side of the Ottawa River
(R2 = 0.79). When just
considering the water samples collected at Petrie Island Beach on the Ottawa
River side of the beach (excluding the Baie des Sables transect), the profile
of detection of the human Bacteroides
DNA marker at the beach was identical to that of station 440.2 (detection of
the marker on the same eight sampling days out of 18 weekly samples). An exploratory cluster analysis was
performed on the human Bacteroides
data from sampling locations at Petrie Island Beach and all upstream sampling
locations at Ottawa River transects and tributaries. This analysis found that most sampling locations upstream
of the municipal wastewater plant outfalls, as well as the tributary locations,
fell outside of the main cluster containing the beach ankle and chest depth
sampling locations at Petrie Island Beach (Figure 14). The tributary most similar to the beach
locations cluster was Bilberry Creek.
Although this is only an exploratory method of examining the data, the
cluster analysis results suggest that occurrence of human fecal pollution at
Petrie Island Beach is more closely associated with fecal pollution sources on
the Ontario side of the River than the Quebec side.
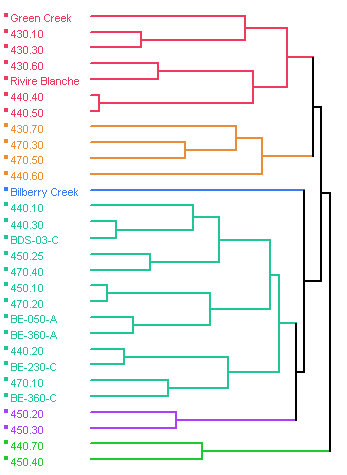
Figure 14. Dendrogram of cluster analysis performed on human Bacteroides DNA marker data collected for sampling locations at Petrie Island Beach and upstream stations on Ottawa River transects and tributaries in 2007. Distances shown by the dendrogram above represent similarity between individual sites for the occurrence of the human Bacteroides DNA marker over the dates sampled. Where sites are clustered in the same group, the occurrence was most similar. Some missing data precluded the inclusion of all sampling locations. Petrie Island Beach sampling locations generally clustered most similarly to Ottawa River sampling locations on the Ontario side of transects (e.g. 440.10, 450.10, and 470.10). They cluster less similarly to sampling locations on the transect upstream from the wastewater plant outfalls (e.g. 430.10), and sampling locations on the Quebec side of the Ottawa River (e.g. 450.40).
Water quality at Petrie Island Beach was generally good in 2007, and there were relatively few beach postings by the City of Ottawa over the bathing season. However, there was evidence of fecal contamination at the beach from both bird and human fecal sources. Bird droppings were numerous along Baie des Sables at times, although the potential for human health risks from these droppings remains uncertain (Edge and Hill, 2007). The detection of the human Bacteroides DNA marker at Petrie Island Beach, along with observations of floatables on the beach at times, is indicative of the impacts of human fecal pollution. Such impacts may present human health risks from the occurrence of waterborne pathogens like Cryptosporidium and Giardia (Chauret et al. 1999) and enteric viruses (Raphael et al. 1985) that are likely to occur to varying degrees in untreated human sewage and the final effluents of municipal wastewater treatment plants. The occurrence of these non-bacterial pathogens may not be entirely predictable using the indicator bacterium E. coli, particularly downstream from fecal pollution sources like chlorinated wastewater effluents.
The impacts of human fecal pollution at Petrie Island Beach were sporadic in 2007 (a relatively dry bathing season), and were mostly associated with rain events. It will be important to better understand when these impacts are likely to occur in the future, and at a minimum, a no swimming rain rule would seem appropriate. The source of this human fecal contamination appeared to be largely restricted to fecal pollution sources on the Ontario side of the Ottawa River in 2007. It will be important to monitor water quality in the future at Bilberry Creek and downstream of the outfall of the City of Ottawa municipal wastewater treatment plant (ROPEC). Close communication between operators of the ROPEC wastewater treatment plant and public health officials overseeing Petrie Island Beach would be needed. The ROPEC plant will need to maintain effective pathogen reduction capabilities, minimize bypass events, and carefully monitor the variability of microbial water quality in the plant’s final effluent to ensure it does not present human health risks at Petrie Island Beach. Bypass events, or changes affecting the normal operation of the plant should be quickly communicated to public health officials. It is important to recognize that there are limitations to using a bacterial indicator like E. coli for making water quality decisions in a riverine setting downstream from a chlorinated outfall of a large municipal wastewater treatment plant. Additional research would be required to better define and remediate the human fecal pollution sources having the most significant impact on Petrie Island Beach.
Many thanks to all the support and sampling assistance provided by Krista Kreling, Mark Macey, and Debbie MacLennan from the City of Ottawa. We also wish to thank the assistance of Sean Abeysekera, Aaron Katz, Gloria Hsiao, Trang Vu, Christine Windsor, Yue Ning Wong, and Dr. Abdel El-Shaarawi from the National Water Research Institute, Environment Canada. Project funding was provided by the City of Ottawa.
Bernhard, A.E., and K.G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Applied and Environmental Microbiology 66: 4571-4574.
Bower, P.A., C.O. Scopel, E.T. Jensen, M.M. Depas, and S.L. McLellan. 2005. Detection of genetic markers of fecal indicator bacteria in Lake Michigan and determination of their relationship to Escherichia coli densities using standard microbiological methods. Applied and Environmental Microbiology 71: 8305-8313.
Chauret, C., S. Springthorpe, and S. Sattar. 1999. Fate of Cryptosporidium oocysts, Giardia cysts, and microbial indicators during wastewater treatment and anaerobic sludge digestion. Canadian Journal of Microbiology 45: 257-262.
City of Ottawa. 2005. Petrie Island Beach Monitoring Study (2003-05) Final Report. Water Environment Protection Program, City of Ottawa. 26p.
Edge, T.A., and K.A. Schaefer (Eds.). 2006. Microbial source tracking in aquatic ecosystems: the state of the science and an assessment of needs. National Water Research Institute, Environment Canada, Burlington, Ontario. NWRI Scientific Assessment Report Series No. 7, and Linking Water Science to Policy Workshop Series. 26p.
Edge, T.A., and S. Hill. 2007. Multiple lines of evidence to identify the sources of fecal pollution at a freshwater beach in Hamilton Harbour. Water Research 41: 3585-3594.
Edge, T.A., A. Crowe, S. Hill, P. Seto, and J. Marsalek. 2007. Surveillance for Sources of E. coli at Toronto’s Bluffers Park Beach 2005-06. Water Science & Technology Directorate Technical Note No. AEP-TN07-003. Environment Canada. Burlington, Ontario. 41p.
Field, K.G., and M. Samadpour. 2007. Fecal source tracking, the indicator paradigm, and managing water quality. Water Research 41: 3517-3538.
Fogarty, L.R., S.K. Haack, M.J. Wolcott, and R.L. Whitman. 2003. Abundance and characteristics of the recreational water quality indicator bacteria Escherichia coli and enterococci in gull faeces. Journal of Applied Microbiology 94: 865-878.
Gawler, A.H. et al. 2007. Validation of host-specific Bacteriodales 16S rRNA genes as markers to determine the origin of faecal pollution in Atlantic Rim countries of the European Union. Water Research 41: 3780-3784.
Gould, D.J. and M.R. Fletcher. 1978. Gull droppings and their effects on water quality. Water Research 12: 665-672
Raphael, R.A., S.A. Sattar, and V. S. Springthorpe. 1985. Long-term survival of human rotavirus in raw and treated river water. Canadian Journal of Microbiology 31: 124-128.
Scopel, C.O., J. Harris, and S.L. McLellan. 2006. Influence of nearshore water dynamics and pollution sources on beach monitoring outcomes at two adjacent Lake Michigan beaches. Journal of Great Lakes Research 32: 543-552.
U.S. EPA. 2005. Microbial source tracking guide. EPA/600-R-05-064. Office of Research and Development. United States Environmental Protection Agency, Cincinnati, Ohio. 150p.
Whitman, R.L., and M.B. Nevers. 2003. Foreshore sand as a source of Escherichia coli in nearshore water of a Lake Michigan beach. Applied and Environmental Microbiology 69: 5555-5562.